explore the human phospho-reactome and actionable tumor kinome
By curating a large number of public databases, PhosphoAtlas created a comprehensive map of human kinase circuits that can be globally interrogated to identify mutated signaling nodes in cancers and to select actionable targets for therapy.
To download the PhosphoAtlas database files, please fill out our registration form.
For technical assistance, please contact us at [email protected].
Links to article: PubMed, Cancer Research, PDF
Summary
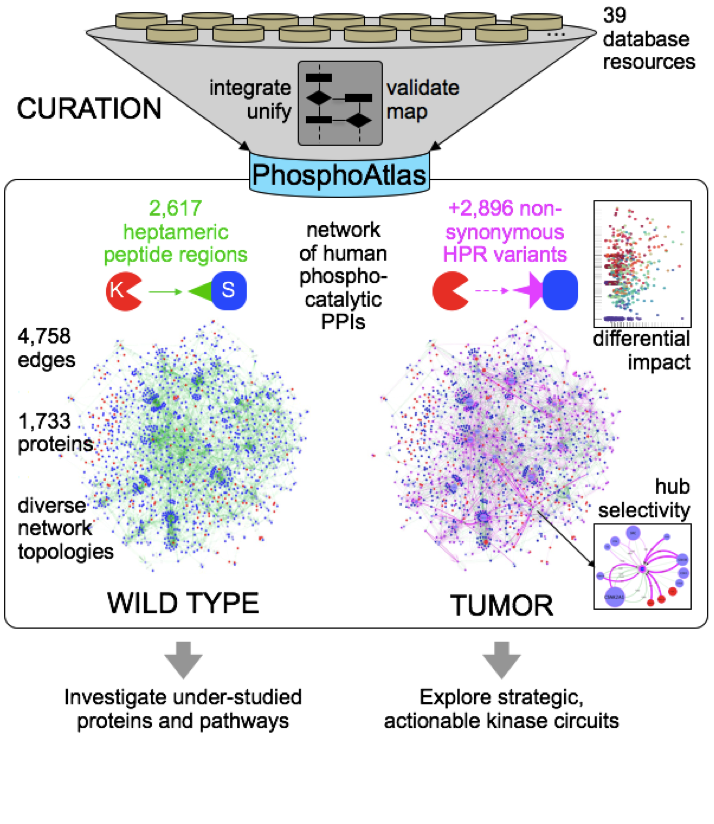
Kinase inhibitors are used widely to treat various cancers, but adaptive reprogramming of kinase cascades and activation of feedback loop mechanisms often contribute to therapeutic resistance. Determining comprehensive, accurate maps of kinase circuits may therefore help elucidate mechanisms of response and resistance to kinase inhibitor therapies. In this study, we identified and validated phosphorylatable target sites agnostically established from any human cell or tissue type to generate PhosphoAtlas, a map of 1,733 functionally interconnected proteins comprising the human phospho-reactome. A systematic curation approach was used to distill protein phosphorylation data cross-referenced from 38 public resources. We demonstrated how a catalog of 2,617 stringently verified heptameric peptide regions (HPR) at the catalytic interface of kinases and substrates could expose mutations that recurrently perturb specific phospho-hubs. The in silico mapping of 2,896 nonsynonymous tumor variants identified across thousands of tumor tissues, also revealed that normal and aberrant catalytic interactions co-occur frequently, showing how tumors systematically hijack, as well as spare, particular sub-networks. Overall, our work provides an important new resource for interrogating the human tumor kinome to strategically identify therapeutically actionable kinase networks which drive tumorigenesis.
Reference
Olow A*, Chen ZZ*, Niedner RH, Wolf DM, Yau C Pankov A, Lee EPR, Brown-Swigart L, van ‘t Veer LJ, and Coppé JP. An atlas of the human kinome reveals the impact of the mutational landscape underlying dysregulated phosphorylation cascades in cancer. Cancer Research. 2016
Highlights
- Kinase–substrate networks are diversely connected via 4,758 catalytic interactions
- 52% of the 2,617-peptide regions are affected by non-synonymous tumor mutations
- Cancer phospho-circuits reveal widespread, yet specific, recurrent aberrations
- This network is a roadmap to explore the strategic, actionable kinase-hotspots of cancer
Impact
Exploring experimentally validated phosphorylation circuits remains a major challenge due to the abundance of available knowledge. Our computational study curated 39 public databases to systematically verify, integrate and map human kinase circuits. The diversity of network topologies and differential mutational susceptibilities of target regions revealed the hyper-variable phospho-hubs of cancer.
Access
Access to files/database is granted upon signing the licensing agreement and having the registration confirmed. For-profit entities wishing to access the database are invited to contact UCSF’s Office of Innovation, Technology and Alliances to discuss potential licensing opportunities. Please email Gemma Rooney and reference the UC Case No. SF2016-209. For scientific inquiries and collaborations, please register and email Jean-Philippe Coppé.