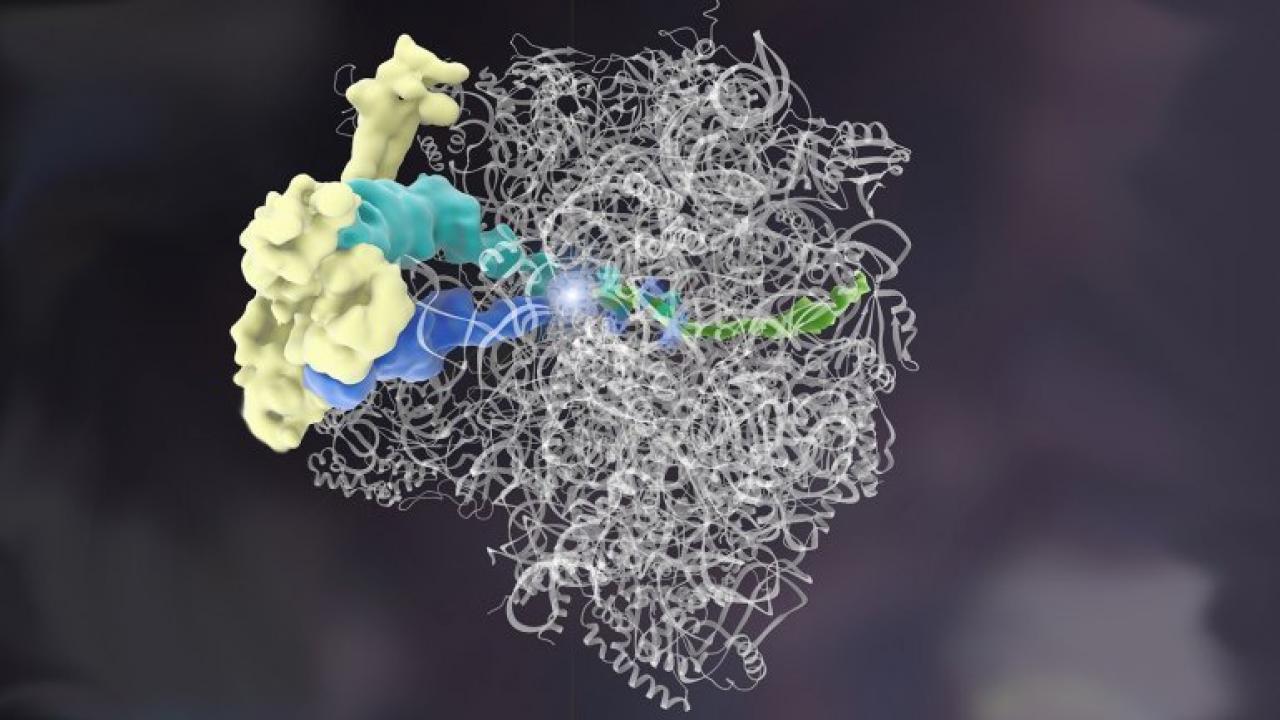
Complex, floppy molecules like this ribosome quality control complex were hard to see at the atomic level because their structures fell apart when researchers tried to prepare the sample for other visualizing techniques. UCSF researcher Adam Frost, PhD, was able to visualize it at last using a new technique. Artistic rendering by Janet Iwasa, PhD
One of the more famous images in biology is known as "Photo 51," an image of DNA that chemist Rosalind Franklin and Raymond Gosling created in 1952 by shooting X-rays through fibers of DNA and analyzing the patterns they left behind on film.
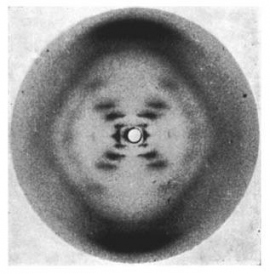
X-ray diffraction image of the double helix structure of the DNA molecule, taken in 1952. The image, famously known as "Photo 51," led to greater understanding of DNA and gave rise to the field of molecular biology. Image by Raymond Gosling/King's College London
That image provided Nobel Prize winners James Watson and Francis Crick with crucial information they needed to determine the 3D structure of DNA – knowledge that ultimately gave rise to molecular biology.
For much of the 20th century, this method of analyzing X-ray diffraction patterns has been the standard for determining the 3D structures of proteins, generally using a technique known as X-ray crystallography. But this method relies on getting proteins to pack tightly together to form uniform crystals, which is notoriously difficult, especially when it comes to the floppy, dynamic proteins that live in cell membranes.
Yet biologists are particularly interested in these membrane proteins because the membrane is the cell’s dock, its security checkpoint, its mailbox. From sugars to viral invaders to nerve impulses, all cellular traffic passes through the cell membrane, and likely interacts with the proteins embedded there, such as neurotransmitter and hormone receptors.
To understand all that transpires at the membrane, biologists need to zoom in to the atomic level. The proteins’ resistance to crystallization, therefore, left scientists in a bind.
Now, after decades of challenges, scientists have developed an imaging technique that allows them to visualize these tiny membrane proteins thousands of times smaller than the width of a human hair – and to do so without first having to form crystals.
Revisualizing a Classic Technique
Known as single-particle cryo-electron microscopy, or cryo-EM, the technique had largely been written off as useless for determining the structure of very small proteins. But technological refinements made over the past several years at UC San Francisco and other institutions have made cryo-EM a key tool in structural biology.

Yifan Cheng, PhD, discusses tweaks to the software that builds the 3D structure from the cryo-electron microscopy images with postdoc Jean-Paul Armache. Photo by Elisabeth Fall

David Agard, PhD, in his office at UCSF Mission Bay
“Cryo-EM has made this marked jump in the last three or four years in terms of resolution, so that we can go from just seeing the general outline of a protein, what people used to refer to as ‘blob-ology,’ and now see, actually, the atomic structure,” said David Julius, PhD, professor and chair of UCSF’s Department of Physiology.
Julius collaborated with Yifan Cheng, PhD, associate professor of biochemistry and biophysics, to use cryo-EM to visualize the structure of the body’s receptors that sense the spiciness of chili peppers and, in work reported last month, wasabi. These receptors are also involved in how the body signals pain – and the findings are already being used to test new pain drugs that bind to these receptors.
“By being able to really determine a molecule’s atomic structure, you can understand how it works in detail, you can make connections between the biology and chemistry, and it also provides capability for drug discovery and design,” said David Agard, PhD, professor of biophysics and biochemistry and a Howard Hughes Medical Institute (HHMI) investigator, who worked with Cheng to create better cameras and software for cryo-EM.
“So it’s really a huge change in what’s possible.”